







Center for Molecular Medicine and Genetics
Wayne State University School of Medicine
540 E. Canfield
Detroit, MI 48201

|
|
| |
|

|
 |
 |
Introduction
Our research has centered on two related themes. One is concerned with the development and application of high throughput technologies for studying gene function (Functional Genomics and Systems biology). The other is concerned with understanding specific cyclin regulatory networks.
Functional Genomics and Systems Biology 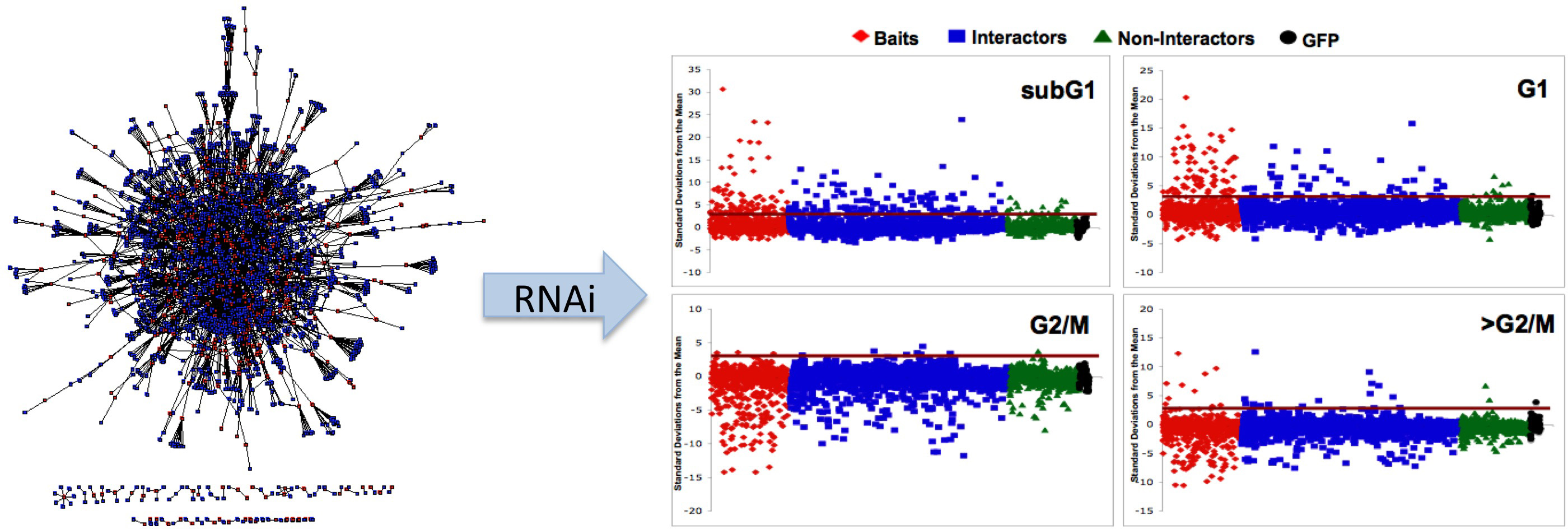
Most of the work in this area has focused on mapping protein-protein interactions (e.g., see Stanyon and Finley, 2000; Zhong et al., 2003; Giot, 2003; Stanyon et al., 2004; Parrish et al., 2007). A long running project was construct and characterize a proteome-wide interaction map for Drosophila. This project entailed conducting high throughput screens, developing computational tools to analyze interaction data, and sharing the interaction data and analysis tools with the public via a web-accessible database (www.DroIDb.org). We have also been concerned with figuring out how to use the flood of functional genomic data from our own and other studies to create systems-wide models of biological processes. In addition to the public database and network analysis interfaces (Murali et al., 2011; Pacifico et al., 2006; Yu et al., 2008), the lab has developed computational methods to analyze interaction networks (Murali et al., 2014), to predict new protein interactions (Yu et al., 2005), to score protein-protein interactions (Yu and Finley, 2009), to expand interaction maps (Schwartz et al., 2009), and to analyze disease gene networks (Amin et al., 2010). We developed methods to use interaction data to understand gene function and regulatory networks, including interaction network-guided RNAi screens to identify new cell cycle regulators and pathways (Guest et al., 2011).
- More information on the Drosophila protein interaction map project
Cyclin Regulalatory Networks
We are interested in understanding how regulatory networks control cell division and survival during development and in diseases such as cancer. Our approach has been to study novel cyclin proteins with sequence similarity to known cell cycle regulators. Students in the lab have studied two poorly characterized but highly conserved cyclins (Cyclin J and Cyclin Y) using Drosophila genetics (Kolonin, 2000; Liu, 2010; Liu et al., 2010; Atikukke et al., 2014). The lab has also constructed and published an extensive protein interaction map centered on cell cycle regulators (Stanyon, et al., 2004). This work highlights a bridge between the two research areas of our lab. Researchers who study specific cyclin proteins do so in the context of gene and protein interaction networks. These networks provide clues about the functions of the novel regulators and can be used to guide experiments that place the novel proteins into regulatory pathways.
Past Projects
Bacterial Pathogens - Protein interaciton maps and clone sets for bacterial pathogens, including C. jejuni, B. anthracis, and S. pneumonia.
A dengue - host protein interaciton map. Identification of interactions between dengue viral proteins and dengue host (human and mosquito) proteins. Mairiang, et al., 2013
Peptide aptamers - Protein networks provide a good starting point to study the functions of individual proteins and regulatory pathways. However, additional approaches are needed to verify interactions and to test the actual functions of particular interactions in vivo. We set out to adress this need by using yeast two-hybrid methods to identify peptides that bind specifically to target proteins and disrupt their activity or interactions with other proteins. We then expressed these so-called peptide 'aptamers' in vivo and examined the resulting phenotypes (e.g., see Kolonin 1998; Kolonin 2000, Soans et al., 2005)
|
| |
|
 |
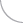 |
 |
 |
 |
|
|
| |
Finley Lab | projects | CMMG home | bookmarks | LabFMdb|
|
|
|

![]()
![]()
