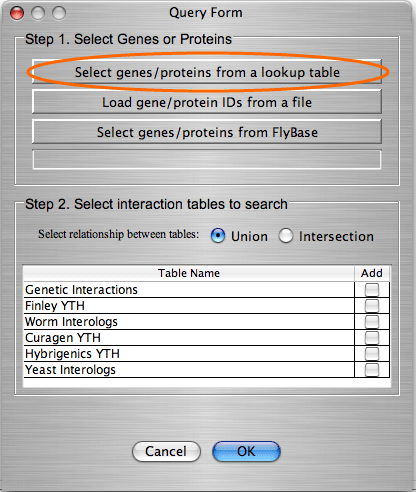
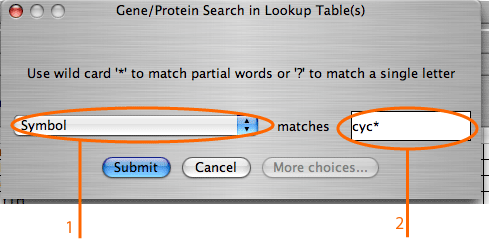
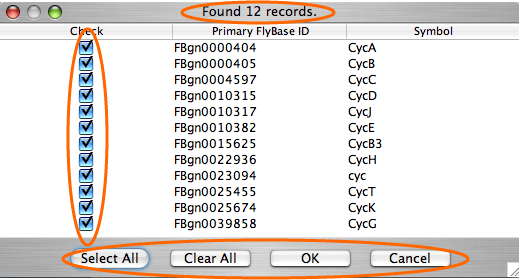
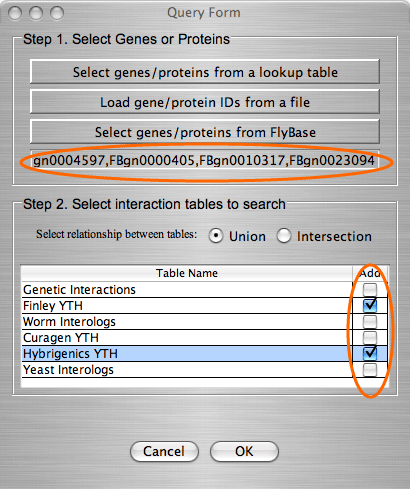
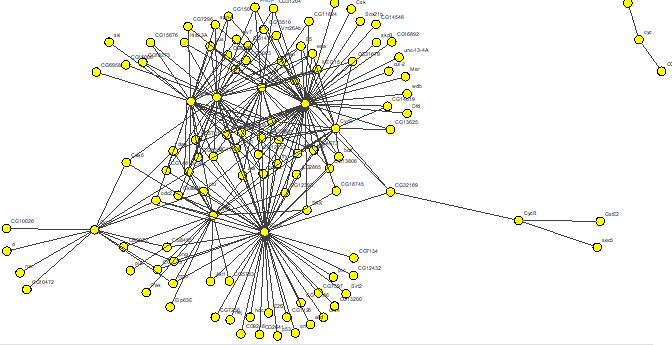
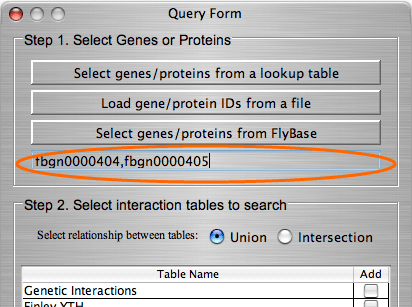
The search by gene/protein attributes is not case sensitive. Queries can be built by exact search term or with wild cards “?” and “*” for a partial terms. In addition to searches on individual attributes, an “Any Field” choice allows all of the fields to be searched simultaneously with the search term entered.
A single gene may have several values for the same attribute; for example, a gene might have several synonyms or several molecular functions. In some databases attributes with multiple values are decomposed into separate tables. In others all values of the same attribute are written in one field, each value delimited by a symbol. IM Browser works with both data organizations. In the first case, when each attribute value stored in a separate row, the user enters the exact value; and in the second case, when several values are stored in one field, user embeds the value into “*”.
For the Drosophila database we included an option for the users to connect directly to Flybase and search its extensive, frequently updated gene and protein information. When deciding whether to search look-up tables or the Flybase database, users should consider the following: searching in look-up tables is much faster than in Flybase database, however look-up tables might contain out-of date information while Flybase is always up to date.
|
![]()
![]()